UPD. For proper tracing how my journey was, I suggest to read all other, in time-related sequence:
- R1a vs. I2a (Jun-2014)
- I-CTS10228 (Jan-2015)
- I-Y4460 (Jun-2017)
- I-B57 (Jan-2018)
- I-Y128456 (Jun-2018)
- I-FGC63213 (Sep-2018)
===
I know YFULL service since maybe 2015, after I started more research in yDNA direction. At that time, I knew that it’s only yDNA tree representation, but I didn’t pay much attention how they do. Later on, 2015-2017, it becomes more popular, and they gave ability to upload DNA raw files to the system and for the dedicated payment they provide more detailed results. I ordered BigY test from FTDNA Aug-2017, but it was delivered only in Nov-2017. And I didn’t have BAM file from FTDNA, which was needed as input file for YFULL. Fortunately, YFULL implemented VCF file analysis, and I was able to upload it and to receive some preliminary results. Jan-2018 I ordered YFULL account based/using VCF (49$), and I knew, that later on I will be able to upgrade with free upload of BAM file. May-2018, FTDNA finally renewed BAM file generation. I requested, and after some time I receive BAM file. Then I uploaded to yfull, so my account upgraded. Jun-2018 got some results from YFULL in Upgrade section. But in fact not so much new info. Anyway, YFULL looks very active service, then have recently published v6.03.41 (Jul-2018), which looks to be following SemVer, and I expect their updates more frequent now. Aug-2019 I received new Big Y-700 results, new BAM file, and I re-uploaded to YFULL, which gave me new ID – YF64515.
Ok, no let’s get more detailed.
Disclaimer: I’m not educated well for the topic, so please take this into consideration, when judging 🙂 Here is short guide tour about YFULL service made by YFULL team. And for more details, u can read their FAQ.
FTDNA vs. YFULL aka build 37 (hg19) vs. build 38 (hg38)
Back to Oct-12-2017 there was message/post from Ted Kandell on Yfull.COM facebook group:
And attached URL. So I started worrying, maybe that is why delay?
On Facebook group there was also another related question (deleted later by admins). Important here is that it’s bright example, how FTDNA is slow comparing to 23andme, who already using Chr 38 standard for ages:
Also, one more important note, for those, who received Build 37 BAM file, and already has uploaded to Yfull:
Then I saw message on FTDNA by Zdenko Marković from I2a FTDNA group:
An important notice for those who have Big Y test results or ordered this test. FTDNA is updating its representation of Big Y result. The most important change is in human genome build. Until now, all your results as well as data in your VCF and BAM files were based on the hg19 human genome representation. As of today they will be displayed in a new hg38 human genome representation. Maybe you noticed that you have „Results Pending“ page when you click on your Big Y section. You will be notified by email once your results are processed and ready. More about hg38 can be found here: https://www.ncbi.nlm.nih.gov/grc/human What does this mean regarding your results? It will not change your haplogroup. It will not affect your place in the haplotree. But it will change the position of your SNPs and your private novel variants. Some new features in your Big Y results representing are also expected. Ray Banks from ISOGG put the comparison table for the known SNPs here: Google Doc YBrowse.org is also updating to hg38.
Stanislaw Plewako from FTDNA Baltic Sea group:
Family Tree DNA have written to Group Administrators about a great changes in classification: ” We’re releasing a big update to Big Y on October 10th …. Once the release is live, we will be recalculating Big Y matches. We anticipate this to take approximately 5-7 days. During this time, you will see a “Results Pending” page when you click on the Big Y section. You will be notified by email once your results are processed and ready. We’ve updated from hg19 to hg38. This is a more accurate representation of the human genome and is the most recent version referenced by the human genome community. Some of the advantages of hg38 are: Better mapping of NGS data to the proper location Consideration of alternative haplotypes across the genome”
Btw, on YFULL, there is such page – HG38 Announcement, where we can read main info.
Ted Kandell Dec-08-2017, on yfull facebook group post comment:
Build 38 however gives some important advantages. (Build 38 was used by FGC for quite a while now, BTW.) It’s a much more accurate Y Reference Sequence, and therefore some regions of false Y sequence are eliminated, and some newly identified regions have been added. There are also some rearrangements of the existing Build 37 sequences. Therefore, when these “reads” (short fragments) are aligned – matched up to the Reference Sequence – they are more likely to “fit’ properly because the Reference Sequence more closely reflects an actual Y chromosome sequence. (A sequence however coming from at least *two* people in very different haplogroups.) Also, some reads may “fit” (be aligned to) some newly added Y sequences, so these reads that were formerly tossed out can now be used. The “false” Y regions also go away too, so there *should* be fewer “junk” SNPs, particularly in the Big Y. However, this doesn’t seem to be the case with the Build 38 Big Y itself. Since Build 38 was available in 2013, FTDNA should have been using Build 38 all along, and not doing so was just carelessness!
So due to delay on FTDNA with BAM file generation, it’s kinda delay with proceed to YFULL upload. But, Jan-13-2017 there was nice update on Facebook yfull.com group page:
I uploaded/requested/ordered YFULL VCF analysis Jan-13-2018, and I hoped that at least those 50% of data can be analyzed first, before FTDNA continue BAM file generation, which happened in May-2018, and I had BAM file, so my final upgrade to BAM was for free on YFULL.
Profile/Account ID
YFULL system assigns ID for every profile after upload. I have YF11414, and now, Y4460 tree updated, with information about me:
Note: in blue color, I added information about closest IDs/profiles/neighbors, and I contacted them already.
v7.06.00 (Jul-2019):
v 7.07.00 (with both my id: YF11414 and YF64515):
Nov-2019, released Chart View:
SNPs
YFULL grouped information about SNPs under one section/menu with different sub menus.
Known SNP – a SNP that has been discovered and named by YFull or by another organization or person. There is a list of organizations and persons at the end of the Positive, Ambiguous and No call tab. Positive SNP – a SNP with a mutation. Also called a derived SNP and a SNP variant. The mutations in a customer’s raw data sample (BAM file) establish the customer’s genetic path from the deep past towards the present. Negative SNP – a SNP without a mutation. Also called an ancestral SNP. About 95% of a customer’s SNPs are negative.
Haplogroup and SNPs
As expected, after analysis finished, I have ability to see available SNPs similar to my, and list of people who matches my DNA also. In FTDNA system, based on latest information from Big Y-500 test results page I have 127814 Known SNPs. When I uploaded VCF, list of Known SNPs was smaller (114739) and now, after BAM upload YFULL shows more – 130789. From which, 1996 positive, 12 ambiguous and 2785 with “no cal position” (where B57 and Y3106 listed). When I download CSV file I can see all details. It shows my Y-haplogroup (Y4460), Hg variants (Y4460* and Y3106*) and Terminal SNP (Y4460).
Positive SNP is interesting list, because it shows me, on which level, on which branches, I’m positive and in which position (which SNP). For example BY30348 shows “on level I-Y4460”, but when I search BY30348 from main page, it gives me multiple results of “positiveness” for other I-Y**** SNPs:
Another example with PF3686:
Those Ambiguous SNPs are very interesting, because in fact it shows, that despite the fact we have different haplogroups like R1a, I2a, G, N, E, etc, there might be mutation, in the same place of chromosome Y for all people, and it then kinda shared/common SNP. Not sure how it works on biological level, but this we definitely discovery for me.
For example, SNP Z1950, which is ambiguous for me, in fact belongs to N hg.

Or Y113, which “kinda” belongs to many haplogroups:
Or Y510_1:
That is why there is no dedicated URLs https://yfull.com/tree/I-Y113 or https://yfull.com/tree/I-Y510_1 – these are not valid URLs.
FAQ: No Call – for a specific SNP or ChrY position in a sample, the YFull analysis did not detect any reads or other information.
As I guess, “No call” section/tab is based on all known SNPs, which are positive for someone else in database, but not found in my DNA at all. It’s odd, considering YSEQ.net gives me B57+ and neither FTDNA nor YFULL shows it at all.
This is my main question for genetics research in 2018.
Novel SNPs
FTDNA system shows my 12 Unnamed results/entries, which are about positions in Chromosome Y, without defined name. Zdenko Markowic from FTDNA also mentioned, that my DNA results show 19 private novel variants.
YFULL FAQ:
- Private SNP – A SNP in a sample in the YFull database is considered private until it has been matched in another YFull database sample with the same “localization”.
- Novel SNP – One type of Private SNP. See FAQ Definitions for Private SNP and YFSxxxxxx.
- YFSxxxxxx – The series of SNPs used to designate YFull’s Novel SNPs that have not yet been added to the Yxxxxxx series by YFull. Sometimes called “YFull singles”.
- YFCxxxxxx – The series of SNPs used to designate SNPs being considered by YFull for naming and inclusion in the YTree. Sometimes called “YFull Candidates.”
So I have 10 best qual Novel SNPs and 5 Acceptable qual SNPs.
Some of Novel SNPs are marked with char G, which is about gene. For more details SNPedia.com will help. Or Promethease report.
Age Estimation
This is new after BAM upload. Age estimation is always important and interesting.
SNPs split in 4 groups, depends on known or novel and if it matters for estimation or not. Snapshot for Jul-2018. And here is the latest (Sep-2018):
Note: Jul-2018 it was 2300 ybp. and now Sep-2018, it’s around 2100 ybp, when first Y4460 mutation appears in male person Y chromosome.
- YF11414 is my account/profile ID, and if this goes somehow unique in future, there will be SNP with my name.
- Y4298 / FGC8449 is shown in I2a haplogroup and Q haplogroup also:

RAW data & SNPs Upgrades
Full statistics
So, using my BAM file, YFULL is able to analyze more details, here is snapshot:
- This information appears ONLY after BAM file upload. Before that, using VCF file, YFULL didn’t show me mtData. But in fact, now there is not much info. Not sure at all, if BAM contains mtDNA data, I kinda doubt. Need to research more.
“Browse raw data” & “Check SNPs”
YFULL gives ability to search/look my raw data using chromosome position.
FTDNA and YSEQ prove, that I’m Y4460+, and looking to YFULL info about Y4460 it proves one more time:
It shows, that YFULL system recognize/remembers hg19 position with number “9028830” but also, new hg38 position “9191221” is already in use. Known SNP in this position is Y4460, and it’s mutation from G to A, which I have also, and this is exactly how it process I’m positive. There is also information from YBROWSE service, which shows more details about SNP.
From FTDNA, even after BigY test, even after I have BAM file, it’s still not clear about B57 and Y3106 for me. YFULL says “no call”.
Anyway, I know, that B57(positive T+ for me based on yseq.net) had a hg19 position 17,061,171 and now it has hg38 position 14,949,291, so I enter this number 14949291 and I can see more details:
 YFULL shows, that there is known SNP B57 C => T, and I am of such. But, when I “Check SNPs” directly, it shows me, that BAM file doesn’t have information about B57:
YFULL shows, that there is known SNP B57 C => T, and I am of such. But, when I “Check SNPs” directly, it shows me, that BAM file doesn’t have information about B57:
And BAM viewer is also empty:
The same situation with Y3106 – “no call position”.
UPD Sep-22-2018. According FTDNA, based on recent Big Y results of new account from Lithuania, FTDNA now can distinguish our shared SNPs in dedicated group/branch. So far on i2aproject blog, it’s named as Y128456, and might be different name in FTDNA and YFULL system soon. But so far, here is info about Y128456 in YFULL:
And also, I’m positive for these SNPs: FGC63213, Y128937, BY51714. Same as that new Lithuanian man.
Still no clue about “no call position” in regards to B57 and Y3106. UPD. Aug-2019 – Positive for both.
VCF vs. BAM viewer
First time, I uploaded VCF file, so after results arrived, I could have used the viewer dedicated to show details about SNP. Here is snapshot with information:

Here is how B57 and Y3106 SNPs/positions looked then:
Yes, it showed, that B57- and Y3106-, and as far as I knew, it’s because of less information from VCF file.
For sure, this viewer might be not relevant anymore, because most of people upload BAM files.
So now, in YFULL system, when I upload BAM file, I can see more details about SNP. For example Y4460 in BAM viewer looks like this:
But B57 looks less information now:
I would expect, at least hint/title. Because I may suspect, that there is no known results for B57 to be positive. But I’m not sure. Same view for Y3106.
Upgrades
So, initially I had VCF file upload, so YFULL did initial analysis, but later on BAM files gave more dat and as result I have more details. And “Upgrades” section shows exact changes. Snapshot for Jul-2018 showed PF6442 the first in list, and now it’s Y510_1 is the first:
- Based on VCF analysis PF6442 was positive for me, now after BAM is “no call”
- Based on VCF analysis Y510_1 was negative for me, but after BAM is positive.
- Based on VCF analysis Y21290 was “no call position’ for me, and now after BAM it’s positive.
- and many other similar changes
- Based on VCF analysis, Y129345 was “Acceptable qual” novel SNP for me, but now after BAM it’s “Best qual” novel SNP.
- Based on VCF analysis, BY148115 was “Low qual” novel SNP for me, and now after BAM it’s “Best qual” novel SNP.
- and many other similar changes.
- old based on VCF is 3427 (2.66%)
- and new based on BAM is 2776 (2.15%)
- Raw Data \ Full statistics it shows 130887 Known SNPs
-
Upgrades \ Statistics for BAM shows 129007 Known SNPs. Again, I assume it should be the same as in raw data.
YFULL vs. FTDNA
Information somehow similar as FTDNA BigY-500 test results. Here is a few SNPs in comparison (statistics collected in early Feb-2018):
FGC30450
- Big Y “Named Variants” tab – YES
- Big Y “Known SNPs” in FTDNA csv file – YES
- YFULL
- VCF: “Novel SNP” in (hg38 pos. 11506442) and marked as private in “Best qual”.
- BAM: “Hg and SNPs” \ Positive.
- Found in search as SNP on CTS10228 level.
PF3827
- yDNA Test Taken – YES
- BigY “Named Variants” tab – YES
- Big Y “Known SNPs” in FTDNA csv file – YES
- YFULL
- VCF: “Known SNPs” on Y4460 level, marked as private.
- BAM: “Known SNPs” on I2 level.
- Found in search as ambiguous.
FGC12093
- BigY “Named Variants” tab – YES
- Big Y “Known SNPs” in FTDNA csv file – YES
- YFULL
- VCF: “Novel SNP” in (hg38 pos. 11504864) and marked as private in “Acceptable qual”
- BAM: “Novel SNP”, Positive, is shown as “Not used for analysis” and “shared” in “Upgrades”\”Novel SNP”
- Not found in search
L138
- BigY “Named Variants” tab – NO
- Big Y “Known SNPs” in FTDNA csv file – NO
- YFULL
- VCF: “Known SNP” – YES (positive for me)
- BAM: “Known SNP” – YES (positive for me on level Y1304)
- Found in search as ambiguous.
Big Y-700 Upgrade
Aug-1029, I Re-uploaded BAM file, and received new ID YF64515.
Results almost the same. There are slight differences, but it doesn’t change my terminal SNP.
Results are almost the same. There are slight differences, but it doesn’t change my terminal SNP.
- For Big Y-500 BAM, Y3113/FGC12083 were ambiguous for me. New Big Y-700 BAM file proves more reliable info.
- For Big Y-500 BAM Y3106 and B57 were “no call”, and Big Y-700 BAM shows as “positive”.
- B57 does have 2 reads, and weight of T is 1.0.
- Y3106 does have 13 reads, but it’s less confident: C(~0.7633), G(~0.2366), error (~0.3347). So it’s shown as positive, but still questionable.
- For Big Y-500 BAM, there were 49 reads, and Big Y-700 BAM proves YFULL to have 46 reads. Less, but seems to be better correctness.
- For Big Y-500 BAM weight for A allele of SNP Y128456 was ~0.9582, which is high enough, but new Big Y-700 BAM gives 1.0 weight. As result no weight of G.
- Same with probability of errors – for old Big Y-500 BAM was ~0.0589, and new Big Y-700 BAM gives 0. Which is obviously better.
- As dependant info from above items, Big Y-500 BAM gives ability YFULL to recognize position data as 47A + 2G (49 reads), and new Big Y-700 BAM as only 46A as position data (all 46 reads).
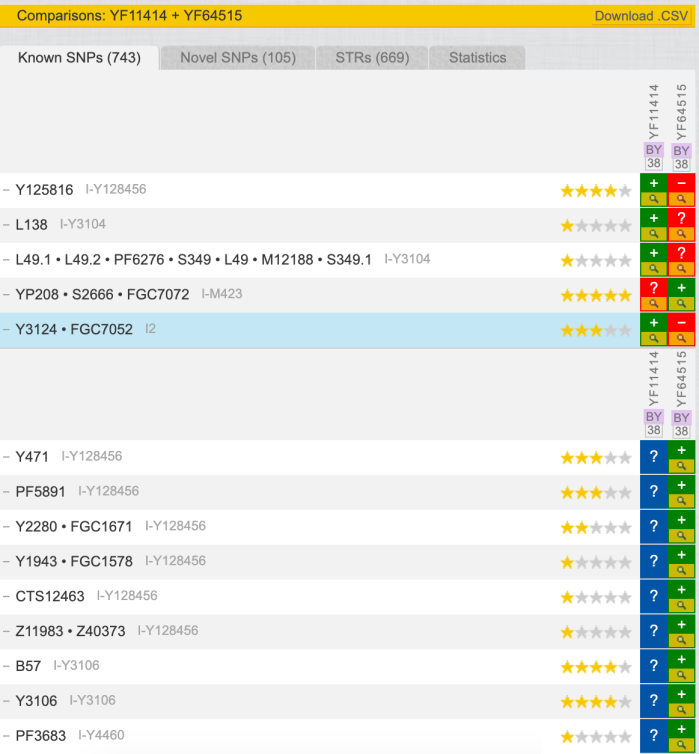
Interesting about comparison between YF11414 and YF64515 data in regards to Known SNPs:
- According old Big Y-500 BAM, I did have Y125816 1 positive read 1A, but for YF64515 it’s gone. Y125816 is also related to Y64438 – child SNP to Y70430, which is Belarus and Greece, so maybe it shows a sign, that algorithm is more granular now, and filter noise and not relevant reads/belongings.
- According old Big Y-500 BAM, I ha “no call” rad of Y471 SNP. And new Big Y-700 BAM gives 1A read. Might be non-relevant, because same read is for yDNA haplorgroup R and O.
- One new read 1A for PF5891 SNP, but again non-relevant, coz it exists also for E, N, P yDNA haplogroups.
How Statistics looks after Big Y-700:
SNPs: Matches
Just a short snapshot – list of SNP matches – people, who does have the same terminal SNP, or at least close with haplogroup.
Notes:
- This list reflects people / accounts which are listed under Y4460 tree branch.
- When VCF analysis done first, the number of shared SNPs, Assumed shared SNPs and All shared SNPs were much bigger. Not sure why – was it a bug or some algorithms changed after I upload BAM file.
Interesting columns are “Shared SNPs” and “Assumed shared SNPs”. Example of my and YF10202 SNPs:
STRs
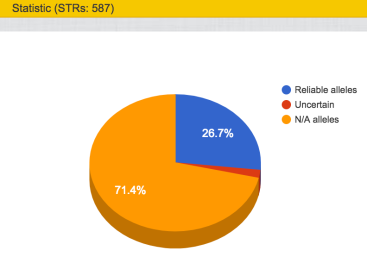
So when VCF analysis done first, there was no STR info. But after BAM file uploaded, I have STR results and even STR matches.
STR Results
Similar data as in FTDNA system, slightly changed in view.
But also some chart representation:
STR Variants
And for example DYS550 (“shared with I-SK1241, I-Y3106, YF10202”) info looks:
STR Matches
So far, matches are far from me (from Y4460). Even close matches are mostly from neighbor SNP S17250. But it’s STR level of matches.
SNPs: YReport
This is dedicated YFULL page, where ALL SNPs shown since Adam till now. It’ huge but shared URL with public. Have a look.
SNPs: My News
SNPs section also shows some News – updates on YFULL system. Not really useull for me so far, but still nice.
Groups
So, as I guessed, L621 group exists. 🙂
 So essentially it’s the same data as in FTDNA system mixed with YFULL system.So far not much new stuff.
So essentially it’s the same data as in FTDNA system mixed with YFULL system.So far not much new stuff.
So, L621 group has nice UI, which shows me/YF11414 with all SNPs in search:
 B57 and Y3106 are kinda of “no call position”.
B57 and Y3106 are kinda of “no call position”.



















































Wasn’t mtDNA in the early BigY bam files but FamilyTreeDNA scrubbed them later on so it wouldn’t compete with their mtDNA offerings?
I don’t know. But for me it’s surprise or simple mismatch of my expectation, that BAM file contained/contains mtDNA info, assuming this all is about yDNA. Anyway, I plan to request YFULL support to explain how BAM file looks when it contains mtDNA info and when it’s not. Maybe it’s different file though.