First time I mentioned about T2T in my article “Why T2T is so important?” in 2022. Based on FTT SNP index I knew that there is now SNP close to my Y-DNA tree and I didn’t order any T2T alignment. But then I thought – why not? Let’s prepare myself for the future, and maybe future T2T re-alignments will bring me some SNPs automatically. So I paid YFULL 23 EUR and Dante Labs 20 EUR.
T2T claims to be a most advanced whole genome sequencing standard so far as of 2022-2024 years.
NCBI
Some info about standards.
16-Nov-2021 => CP086569.1 aka HG002v2 – Homo sapiens isolate NA24385 chromosome Y.
Assembly Method :: T2T assembly pipeline v. 09/13/2021 Assembly Name :: Complete chrX and chrY Genome Representation :: Full Expected Final Version :: No Genome Coverage :: 60.0x Sequencing Technology :: PacBio HiFi; Oxford Nanopore GridION; Oxford Nanopore PromethION
PacBio Circular Consensus Sequencing (CCS) of the human male HG002/NA24385 with 15 kb and 20 kb inserts to support the development of benchmark standards for small and large variants, haplotype phasing, and de novo genome assembly.
04-APR-2022 => CP086569.2 aka hs1
YBrowse says: “Human Y Experimental T2T J1-ZS2712“.
On Jan 24, 2022 this sequence version replaced CP086569.1.
12-APR-2022 => CM034974.1 aka HG01243 – Homo sapiens isolate HG01243 chromosome Y, whole genome shotgun sequence
YBrowse says: “Human Y Experimental T2T R1b-DF27“
Assembly Date :: 10-SEP-2021 Assembly Method :: MaSuRCA v. 3.4.3; Flye v. 2.5; Nextpolish v. 1.3.1 Assembly Name :: hg01243.v3.0 Genome Representation :: Full Expected Final Version :: No Genome Coverage :: 282.0x Sequencing Technology :: Illumina NovaSeq; Oxford Nanopore; PacBio Sequel; PacBio HiFi
YBrowse
YFULL refers to T2T standard “CP086569.2” and YBrowse does have only two:
- CM034974.1 Human Y Experimental T2T R1b-DF27 (aka HG01243)
- CP086569.1 Human Y Experimental T2T J1-ZS2712 (aka hs1)
I assume, YBrowse will upgrade list in future.
FTT SNP index
The SNPs page refers to at least two standards:
- CP086569.1 aka HG002v2.
- CP086569.2 aka hs1.
YFULL
So, Feb-2024 I finally decided to order T2T upgrade. I knew that will not bring me a surprise, but I started seeing that many other Y-DNA researchers constantly write about T2T, mention their new SNP from T2T, YFULL released new YTree version when designates T2T related SNP with (T) suffix. So…
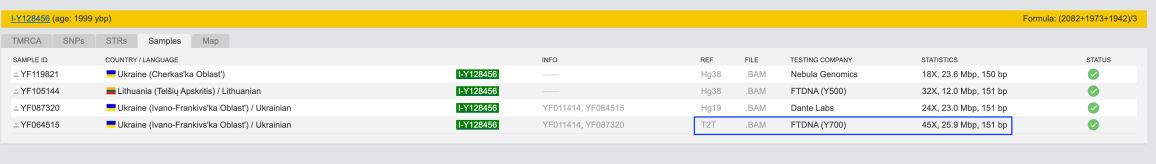
My account which previously was designated as hg38 now after upgrade on Public tree under I-Y128456 is designated as T2T:
Although on Comparison \ Statistics it is still designated as hg38:
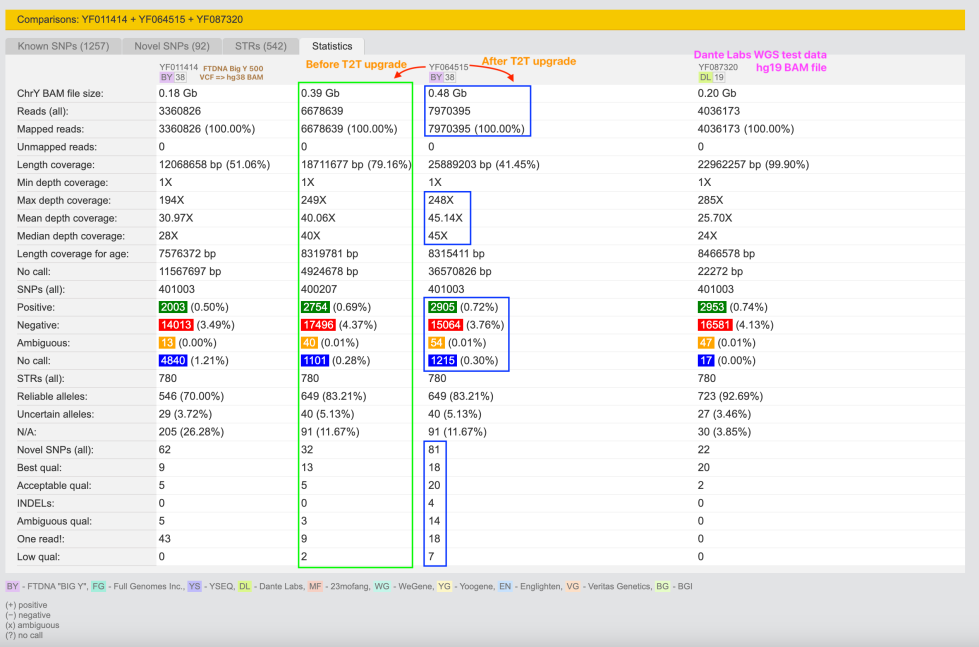
In general, I didn’t expect much, but if I look to YReport comparison I see some slight changes (let’s look to L621 scope only):
Before T2T upgrade:
After T2T upgrade:
On I-L621 there are other (T) SNPs. For me it’s not important on the this level of my research.
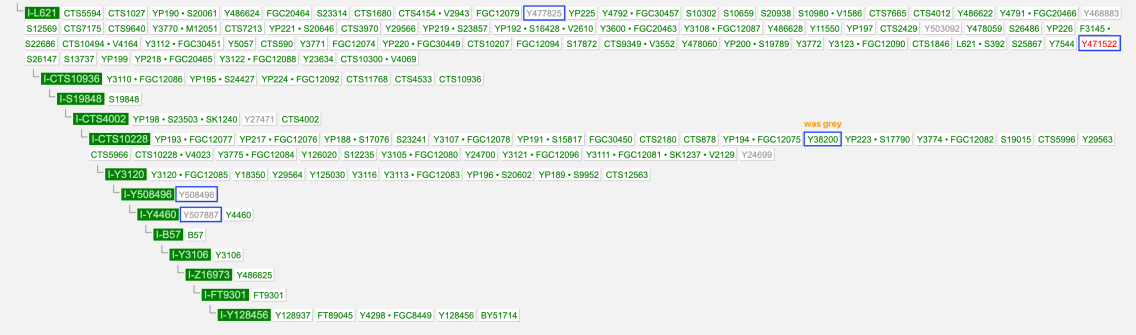
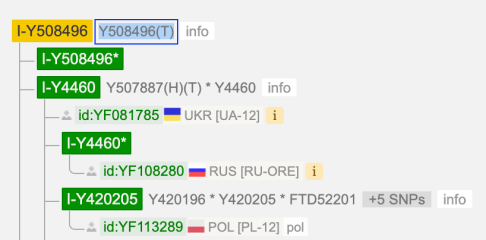
I-S20602 (I-Y3120) \ Y508496
New for me to realise is SNP I-Y508496 which is directly UNDER I-S20602 (I-Y3120) level
(And it seems to be parent SNP for I-Y4460, I-S17250 (I-Y3548) but sibling SNP with I-Y18331 and I-Z17855)
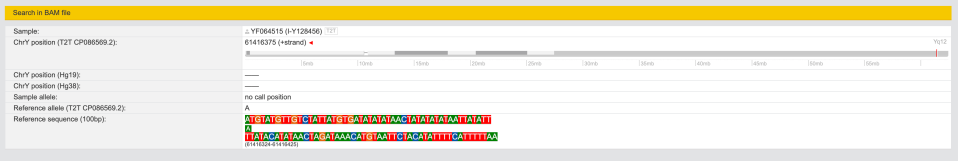
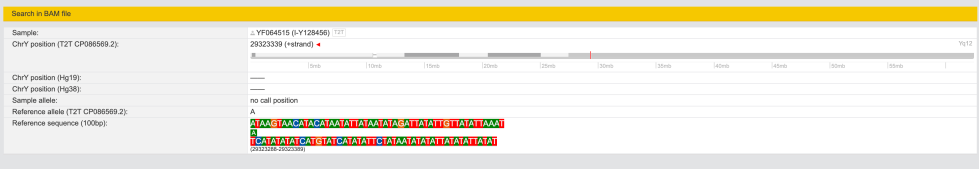
Clicking on left on search icon I get this:
As of Feb-22-2024 it says “no call position” which explains why it’s grey (ambiguous).
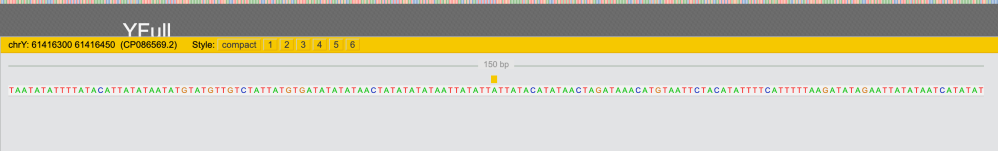
And when I click on “.BAM” icon I now have some data.
chrY: position: 61416300 – 61416450 (CP086569.2)
On Y508496 tree new SNP is designated as “Y508496(T)” (meaning it’s positive for ONLY for T2T):
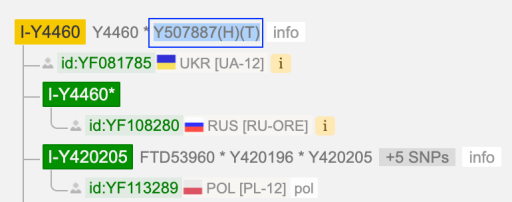
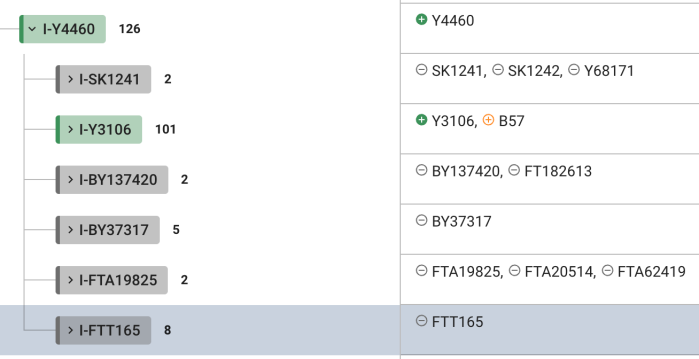
I-S20602 (I-Y3120) \ I-Y4460 ~ Y507887
New for me to realise is SNP I-Y507887(H)(T) which is on the level of I-Y4460.
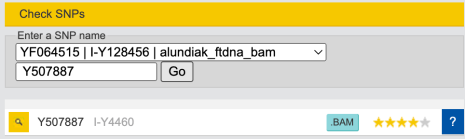
Clicking on left on search icon I get this:
As of Feb-22-2024 it says “no call position” which explains why it’s grey (ambiguous).
And when I click on “.BAM” icon I now have some data.
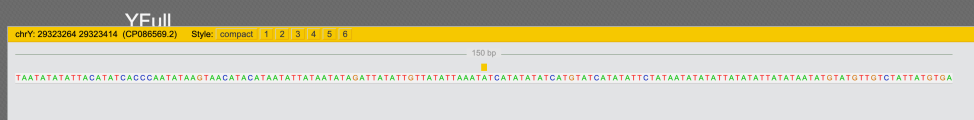
chrY: 29323264 – 29323414 (CP086569.2)
On Y4460 tree new SNP is designated as “Y507887(H)(T)” (meaning it’s positive for hg38 and T2T):
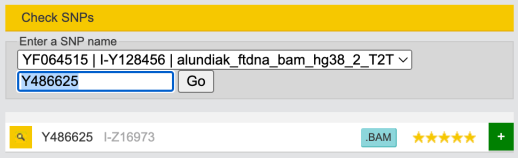
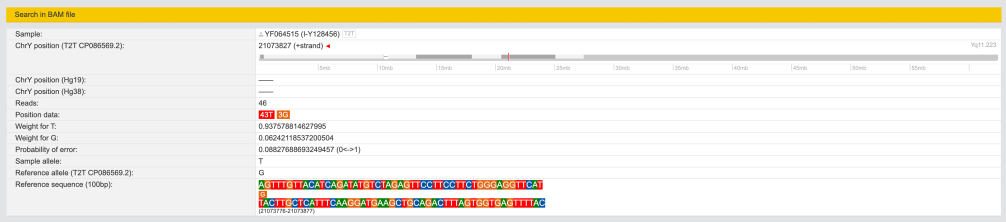
I-S20602 (I-Y3120) \ I-Y4460 \ I-Z16973 ~ Y486625
On I-Z16973 there is another SNP Y486625(T) which I’m positive (which means that sooner or later it will be in my SNPs chain).
Note, that it’s same level as SNP I-FT10545 according to FTDNA nomenclature.
chrY: 21073752 – 21073902 (CP086569.2)
No data on YBrowse yet.
So full path by YFULL SNP nomenclature from I-P37 to my terminal SMP would be:
I-P37 \ I-M423 \ I-FGC41353 \ I-Y3104 \ I-L621 \ I-CTS10936 \ I-S19848 \ I-CTS4002 \ I-CTS10228 \ I-Y3120 \ I-Y508496 \ I-Y4460 \ I-B57 \ I-Y3106 \ I-Z16973 \ I-FT9301
Dante Labs
I ordered T2T alignment because I wanted to know how their order works, 20 EUR wasn’t expensive and I wanted to have ANOTHER T2T aligned BAM file other than from YFULL. Nevertheless there are STILL some doubts about quality of Dante Labs alignment. See details in my previous article – Why T2T is so important?
I didn’t have any emails about finish the process, but after a few days, I simply looked to Dante Labs portal, and I realised I got MORE raw DNA files:
The largest file is BAM (T2T) which is around 97GB. And you better DOWNLOAD it because Dante Labs after some time move it to archive.
UseGalaxy
There is ability to use your *.BAM or *.CRAM file or maybe *.FASTQ file and then upload to UseGalaxy and receive your own FREE T2T aligned *.BAM file.
More detailed in my article here => How to generate T2T BAM file on Galaxy FOR FREE
FTDNA
FTDNA doesn’t have de facto a T2T alignment as a feature to oder, but the Y-DNA tree got updated by some FTT-XYZ SNPs anyway and by having Big Y 700 it’s real, strategical benefit.
For example the closest to me is my parent SNP I-S20602 (I-Y3120) \ I-Y4460 does have its child SNP/subclade SNP FTT165 (which I am negative):
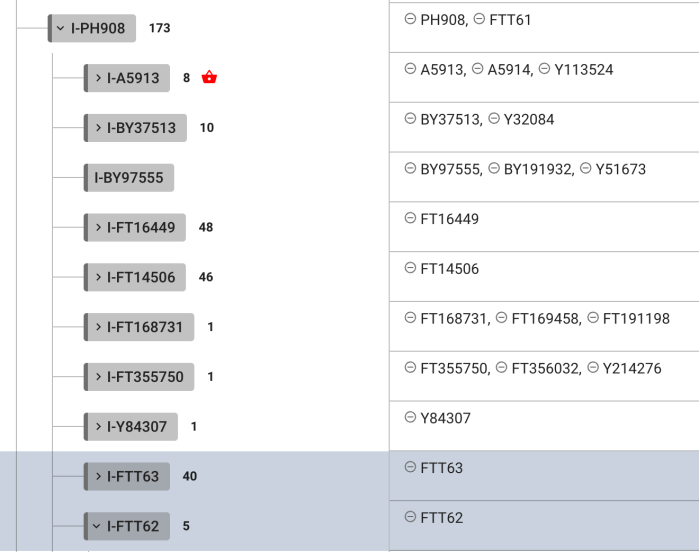
Another close but still far cousin branch I-S20602 (I-Y3120) \ I-S17250 (I-Y3548) \ I-PH908 where under the branch found at least a few: FTT60, FTT61, FTT62, FTT63, FTT65 (all negative for me)
I assume, and hope that in future if any new T2T re-aligned standards will provide new SNPs it will appear on the tree.
I also assume that in nearest future there will be NEW SNP on FTDNA Y-DNA tree which will aggregate Dinaric North (Y4460) and Dinaric South (I-PH908). And YFULL seems to be faster in this direction.
Resources
- Telomere-to-Telomere (T2T) (genome.gov)
- FTT SNP Index (isogg.org)
- Y-DNA_next_generation_sequencing (isogg.org)
- quarTeT: a telomere-to-telomere toolkit for gap-free genome assembly and centromeric repeat identification (ncbi)
- Gapless Telomere to Telomere human genome (T2T-CHM13) now available (ncbi)
- Genome assembly T2T-CHM13v2.0 (ncbi)
- Genome assembly GRCh38.p14 (ncbi)